Academic Publications
Research Sponsors
- National Institute of General Medical Sciences (R01GM127267, P41GM103390, R01GM096049A, P20GM104932)
- National Science Foundation Division of Chemistry (1608685)
- GenNext Technologies, Inc.
- The University of Mississippi
Hydroxyl Radical Protein Footprinting
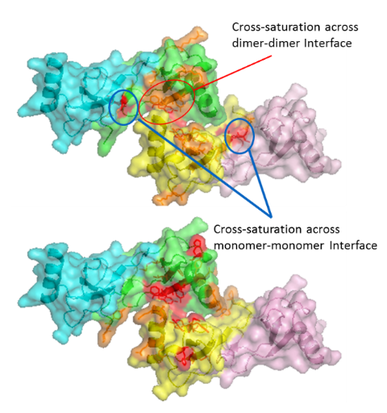
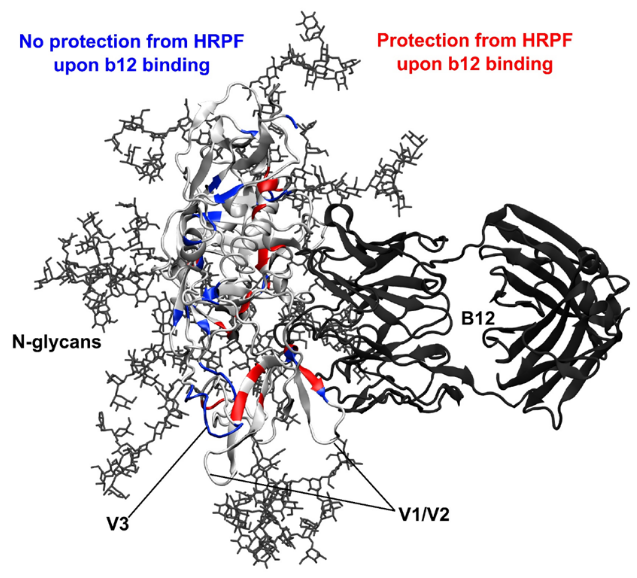
One of the major projects in our group is the development and application of new methods for studying protein structure, the most notable of which is hydroxyl radical protein footprinting. Hydroxyl radical protein footprinting (HRPF) coupled with liquid chromatography-mass spectrometry (LC-MS) has become an increasingly popular structural analysis technique to probe changes in protein topography, offering high labeling efficiency, stable modification products, and a flexible detection scheme. A protein is exposed to a short-lived concentration of hydroxyl radicals freely diffusing in solution, which oxidizes the amino acid side chains of the protein. Amino acid side chains are oxidized at a rate based on a combination of their inherent reactivity and the accessibility to the hydroxyl radical diffusing in solution. By comparing the same protein in two or more conformational states (e.g. ligand-free versus ligand-bound), semi-quantitative measurements of changes in the protein topography can be made. Because of its flexibility and labeling efficiency, HRPF has been successfully used to measure protein folding, protein conformational change, protein-protein interactions, and protein-ligand binding surfaces in a wide variety of proteins.
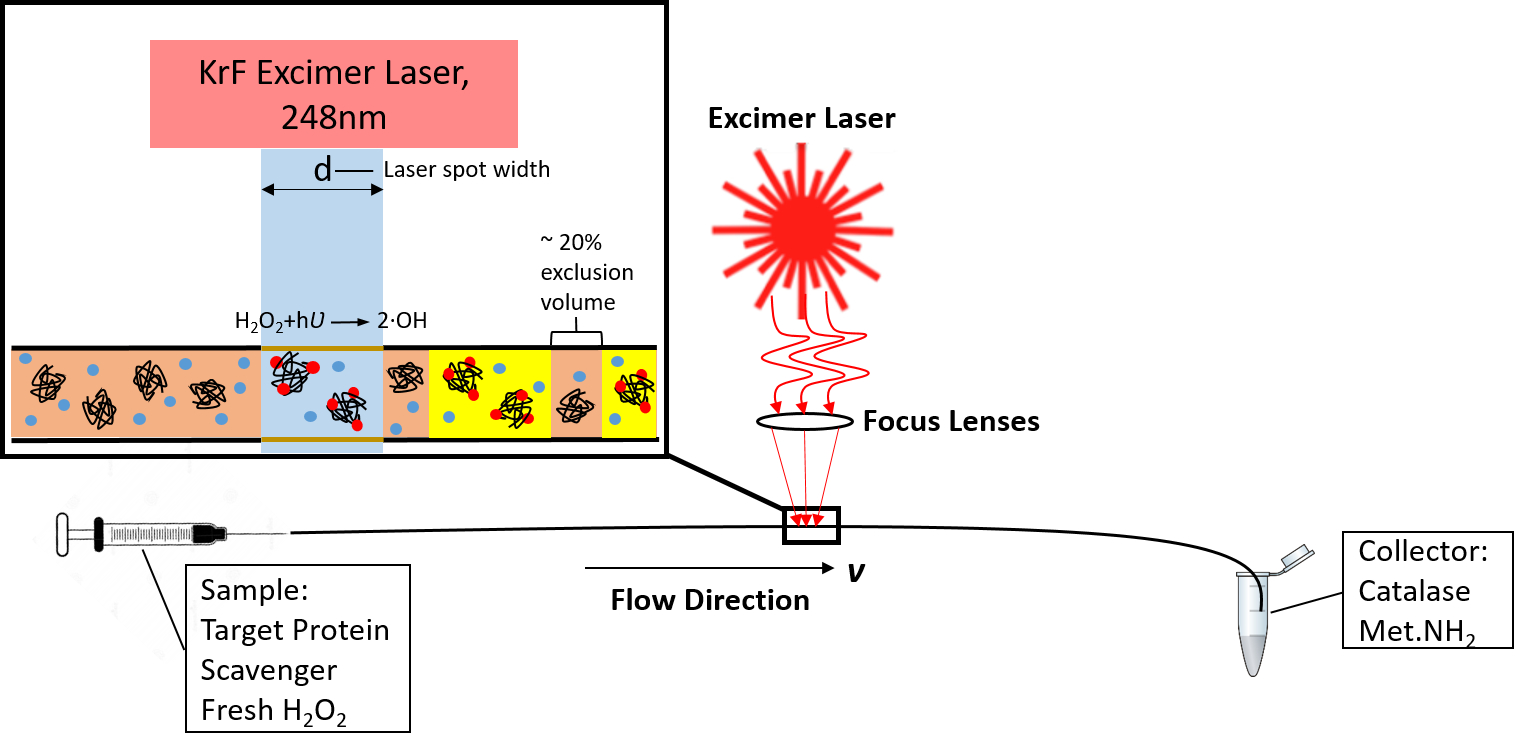
In our group, we have worked to help develop an ultra-fast method for generating these radicals, known as fast photochemical oxidation of proteins (FPOP). FPOP offers superior labeling efficiency compared to other methods, as the lifetime of the labeling process is controlled to complete all protein oxidative labeling faster than the protein can unfold. This allows researchers to probe many more amino acids than prior methods without introducing artifactual data. This heavier labeling extent brings unique analytical challenges with reaction control, measurement, and data interpretation. Our group is at the forefront of developing new technologies and capabilities for FPOP analysis, as well as to make FPOP technology more accessible to new labs looking to enter the field. Our ongoing advancements in FPOP radical dosimetry/actinometry, electron transfer dissociation-based methods for accurately measuring oxidation at single amino acid spatial resolutions, and improved methods for LC-MS/MS acquisition and data analysis look to develop FPOP into a routine technology in the structural biology arsenal. We commonly collaborate with structural biologists to both validate and extend their findings into more complex biological and biochemical systems.
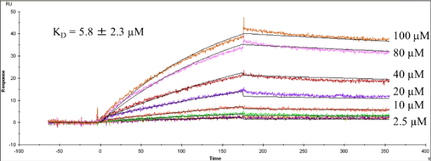
Our group also is active in developing novel applications for HRPF by FPOP, most notably in the field of glycoproteins, proteoglycans, dynamic conformational systems such as protein oligomerization and intrinsically disordered proteins, and protein-carbohydrate interactions, where the extreme heterogeneity and flexibility of the carbohydrates make traditional structural biology tools challenging to use. We seek to better understand the basic chemistries behind HRPF, with the goal of using HRPF FPOP to generate quantitative measurements of protein biophysical properties. Ultimately, our group seeks to solve an unknown protein structure using a combination of HRPF experimental data and computational modeling.
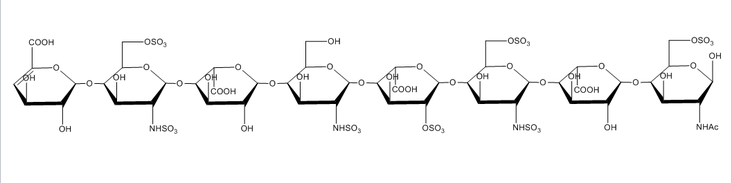
Glycosaminoglycans
Glycosaminoglycans are long unbranched polydisperse polysaccharides consisting of repeating disaccharide units and are found on the cell surface of all mammalian cells. Glycosaminoglycans are involved in a wide variety of cell-cell, cell-protein, and cell-pathogen interactions, and specificity of these interactions are driven by the pattern of modification of the polysaccharides, most notably N- and O-sulfation, N-acetylation, and epimerization of uronic acid residues. Because of the extreme diversity in these modifications, understanding the link between glycosaminoglycan structure and glycosaminoglycan function is a very challenging problem. Due to their almost ubiquitous role in interactions at the cell surface, our group has sought to develop new tools to meet this challenge.
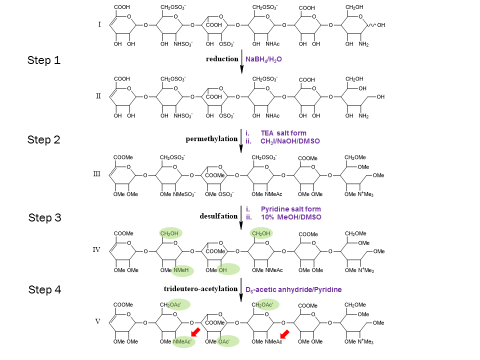
The extreme fragility of the sulfate groups on the glycosaminoglycan oligosaccharides has led us to pursue a chemical derivatization strategy to help us separate and sequence these oligosaccharides. Our strategy seeks to specifically replace all labile sulfates with stable functional groups, protecting all non-sulfated N- and O-groups by permethylation. This results in an oligosaccharide that can be separated by reverse-phase LC, and that yields diagnostic fragments by MS/MS for both sulfate position and uronic acid epimerization. Our group is actively involved in seeking to improve the efficiency of this derivatization chemistry, as well as improve methods for glycosaminoglycan depolymerization and oligosaccharide isolation. Ultimately, we seek to apply our tools to identify specific binding oligosaccharides that mediate various cell surface interactions, with the intention to generate or isolate drug candidates to modulate these interactions for a wide variety of biomedical purposes, including anti-infective agents.